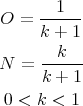
The DNA polymerase makes errors at a low but finite rate that varies depending on the enzyme, condition of the reaction, and the sequence [132]. For example, the errors produced by Taq polymerase are primarily single-base substitutions. That rate can be higher than 10-3 per nucleotides at high Mg2+ and high nucleotide concentrations, and less than 10-6 per nucleotide under other conditions.
During the PCR, amplification products serve as templates for the subsequent cycles. Therefore, sequence changes caused by errors of the DNA polymerase are “inherited”. Since there is no selection based on the functional significance of sequence information, these sequence changes can accumulate. Consequently, a significant percentage of the amplified fragments may carry “mutations”. The percentage of fragments with a correct sequence can be mathematically estimated, assuming a random distribution of errors, and that both mutated and correct sequences are amplified with the same efficiency [133, 134]. If amplification proceeds with an efficiency of k, then the amount of old (O, present at the beginning of the cycle) and new (N, synthesised during the cycle) strands at the end of each cycle have the following relationship:
 |
The probability of producing fragments without error (p) in one cycle of amplification is given by the probability of no-hit in a Poisson distribution.
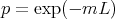
where m is the error rate of the polymerase per nucleotide and L is the length of amplification unit in nucleotides.
Therefore, the fraction of strands with a correct sequence after n cycles, F(n), can be estimated by
![n [1 + k exp(- mL)]n
F (n) = f = ------(k +-1)n------](start_html11x.png)
This estimate may not be appropriate if the reaction is initiated from a very small amount of DNA (e.g., less than 103 molecules), when the timing of occurrence of the first mutation can significantly alter the error rate.
When the errors during PCR are distributed evenly throughout the fragment, no particular mutated sequence constitutes a major subpopulation. In this case the predominant species at each nucleotide position in the product is that of the initial sequence, and direct sequencing of the product is an appropriate method. The situation is different, when PCR products are used as cloning substrates. Each clone originates from a single molecule in the PCR product mixture, and therefore may represent errors occurring during the amplification process.
© 2001 Alexander Binder