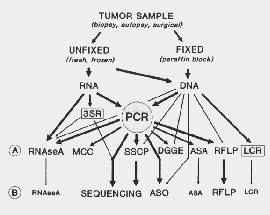
Figure 4.1: Methods for the detection (A) and characterization (B) of point mutations
Due to their subtle nature, the point mutations are the most difficult to detect of all the genetic alterations. Nevertheless, there are quite a few techniques capable of recognizing the presence of single base changes in mammalian genes. A diagram of the methods currently utilized for detection and characterization of single base substitutions, deletions, and insertions is shown in figure fig:mutmeth. In the first row (A) are listed methods for detection of the presence of the mutation. In (B) are the methods that permit characterization of the molecular nature of the mutation. While detection of the presence of a mutation might be sufficient for some diagnostic and prognostic purposes, characterization of the single base substitutions is also important for several reasons.

Figure 4.1: Methods for the detection (A) and characterization (B) of point
mutations
The RNase A mismatch cleavage method is based on the ability of pancreatic ribonuclease to recognize and cleave single base mismatches in RNA:RNA [127] and RNA:DNA heteroduplexes [128]. The method is performed by hybridization of the target sequence to a labeled complementary riboprobe, digestion with RNase A, and analysis of the resistant products by polyacrylamide electrophoresis in denaturing gels. Mutations are detected and localized by the presence and size of the RNA fragments generated by cleavage at the mismatches.
The single strand conformation polymorphism (SSCP ) is another method developed by HAYASHI, SEKIYA, and colleagues [129, 130]. It is based on the differences in the secondary structure of single-strand DNA molecules differing in a single nucleotide, which also is frequently reflected in an alteration of their electrophoretic mobility in nondenaturing gel electrophoresis.
In denaturing gradient gel electrophoresis (DGGE ), the double stranded DNA is subjected to electrophoresis in gel that has an increasing concentration of denaturant along the length of the gel [131]. The fragment melts while traveling through the gel. The melting proceeds in segments, called melting domains, because of the cooperative nature of the denaturation of the double-stranded DNA [132]. When a domain melts, the fragment assumes a branched structure that causes significant retardation of movement. Thus, the position of the fragment in the gel after a certain time of electrophoresis is determined by the history of melting of the fragment that is altered if the sequence is different. The principle of separation in DGGE is such that sequence changes in the melting domain of highest stability cannot be detected, because the fragment no longer has a branched structure when the last domain melts. If, however, a stretch of sequence that serves as an extremely stable domain is attached to one side of the fragment, then mutations at any sites within certain types of sequence context can be detected by DGGE [133]. This extra sequence of extremely high stability can be conveniently attached to the target sequence of PCR by using one primer that has 40 nucleotides of an artificial GC-rich sequence (GC-clamp) extending at its 5'-end. With the use of this clamp, DGGE may be able to detect nearly all possible mutations in any given sequences[134, 135].
Hybridization with radioactively labeled allelic specific oligonucleotides (ASO ) also has been applied to the detection of specific point mutations [136]. The method is based on the differences in the melting temperature of short DNA fragments differing by a single nucleotide. Single point mutations have been also detected by the creation or destruction of restriction fragment length polymorphisms (RFLP).
We successfully established a DGGE protocol for scanning three of the found mutations. Anyway, this method didn't result in the required specificity and therefore a second technique was searched for. As the found point mutations altered three restriction sites of the human 2ar gene, the method of choice was the restriction fragment length polymorphism (RFLP).